Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis.
Fernando G. Osorio*, Axel Rosendahl Huber*, Rurika Oka, Mark Verheul, Sachin H. Patel, Karlijn Hasaart, Lisanne de la Fonteijne, Ignacio Varela, Fernando D. Camargo# and Ruben van Boxtel#
Cell Rep. 2018 Nov 27; 25(9): 2308–2316.e4.
doi: 10.1016/j.celrep.2018.11.014
Abstract
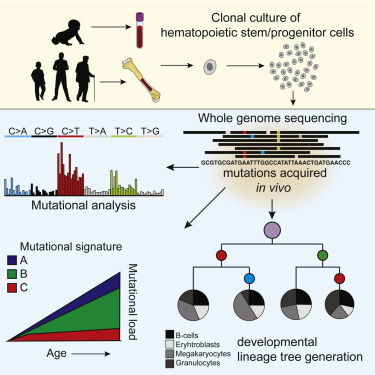
Mutation accumulation during life can contribute to hematopoietic dysfunction; however, the underlying dynamics are unknown. Somatic mutations in blood progenitors can provide insight into the rate and processes underlying this accumulation, as well as the developmental lineage tree and stem cell division numbers. Here, we catalog mutations in the genomes of human-bone-marrow-derived and umbilical-cord-blood-derived hematopoietic stem and progenitor cells (HSPCs). We find that mutations accumulate gradually during life with approximately 14 base substitutions per year. The majority of mutations were acquired after birth and could be explained by the constant activity of various endogenous mutagenic processes, which also explains the mutation load in acute myeloid leukemia (AML). Using these mutations, we construct a developmental lineage tree of human hematopoiesis, revealing a polyclonal architecture and providing evidence that developmental clones exhibit multipotency. Our approach highlights features of human native hematopoiesis and its implications for leukemogenesis.
Keywords: HSC, human hematopoiesis, somatic mutations, mutational processes, leukemia, developmental lineage tree


