Theme 2: Tracking the origin of cancer
DNA is the largest biomolecule in the cells, which unlike other biomolecules is irreplaceable. Consequently, mutations resulting from incorrectly or unrepaired DNA damage will gradually accumulate throughout the life of a cell. The processes causing these mutations leave characteristic patterns in the DNA, such as specific base substitutions in certain nucleotide context (i.e., mutational signatures/footprints). These patterns can serve as a functional readout of mutagenic and/or DNA repair activity. In addition, cells with disadvantageous mutations will be selected against causing a disbalance between damaging and neutral mutations at functionally important loci, such as genes. This phenomenon can be exploited to measure selective pressure. Finally, cells with a more recent common ancestor will share a larger number of somatic mutations than cells that separated early during development, providing means to measure clonal lineages within tissues. Therefore, the complete catalogue of somatic mutations in a genome can serve as an archive containing information on the life history of the cell. By studying mutation accumulation in noncancerous cells of individuals who developed cancer, we aim to determine the mechanisms and rate-limiting steps underlying carcinogenesis.
Strategy 1: Dissecting the etiology of mutational signatures in cancer
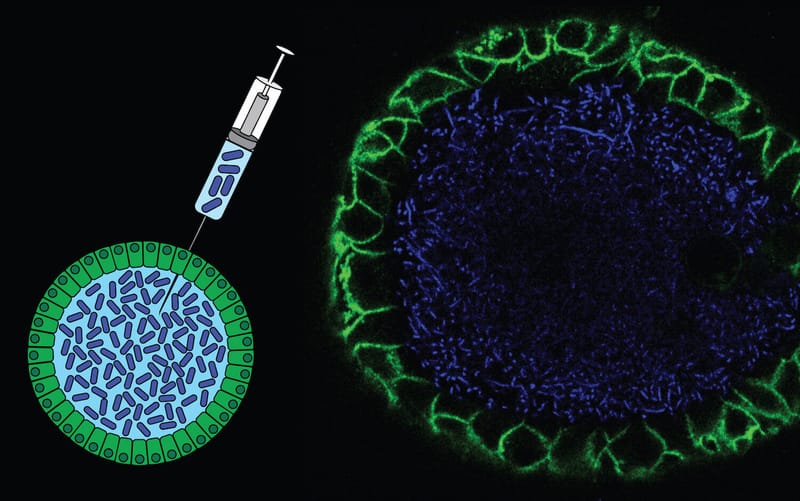
A multitude of mutational signatures have been identified in cancer genomes. However, the mechanisms underlying many signatures remain unknown, which hampers their use in cancer diagnostics. We aim to functionally characterize the etiology of mutational signatures in cancer genomes. To achieve this, we combine experimental and bioinformatics approaches to systematically identify and characterize the processes underlying mutational signatures. For this, we apply CRISPR/Cas9 genome editing or specific carcinogenic exposure in human organoid cultures as well as cell lines. Using this strategy, we discovered a novel signature induced by common gut bacteria that can cause colorectal cancer. Also, we found a signature that can be predictive for genetic predisposition to cancer as a consequence of germline mutations in the DNA repair gene NTHL1.
Publications:
- Drost et al., Science 358:234-238 (2017) PMID: 28912133
- Blokzijl et al., Genome Medicine 10:33 (2018) PMID: 29695279
- Pleguezuelos-Manzano et al., Nature 580:269-273 (2020) PMID: 32106218
Strategy 2: Retrospective lineage tracing of childhood leukemia and lymphoma
Why do children get cancer? For certain cancer types, such as leukemia, young children show a higher incidence compared to young adolescents. This phenomenon represents an apparent paradox, as young cells should have less somatic (oncogenic) mutations than adult cells. Therefore, the etiology of pediatric cancers may differ from cancer in the elderly. We aim to determine the rate-limiting steps underlying the genesis of childhood cancer by studying mutation accumulation and clonal lineages in blood of children with leukemia or lymphoma. For this, we study the processes that contribute to the development of childhood leukemia/lymphoma by comparing somatic mutation patterns between cancerous and patient-matched normal hematopoietic cells. Secondly, we aim to pinpoint the clonal origin of childhood leukemia during the development of the hematopoietic system by retrospective lineage tracing.
Publications:
- Brandsma et al., Blood Cancer Discov DOI: 10.1158/2643-3230.BCD-21-0010


